Areas of Research
Areas of Research for SCCCS
SCCCS is conducting research in the following areas
Microbial Bioremediation of Environmental Pollutants: Environmental pollution is the most significant public and environmental health threat. Due to the growing economy, urbanization and overpopulation growth, we release excess pollutants into the natural ecosystems and destroy their self-cleaning capacity and functionality. Although several methods of pollutant degradation and ecosystem restoration have been practised, microbial-based Bioremediation of pollutants is one of the economical, efficient and feasible options. SCCCS uses the state-of-the-art cutting-edge approach of Monitored Natural Attenuation (MNA), Biostimulation and Bioaugmentation to treat the contaminated soil, water, and landfill leachate using native microbiota of contaminated sites.
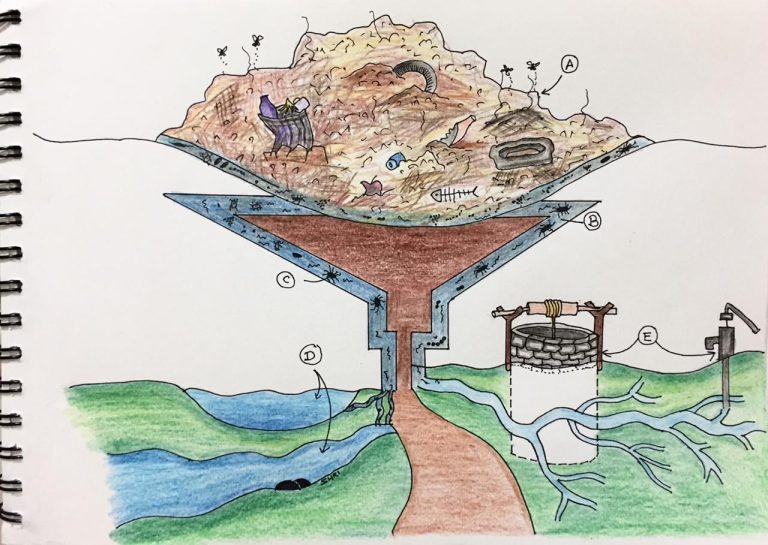
Graphical representation of impact of landfill leachate on environmental and public health. A, landfill or garbage dumping site; B & C, contaminated leachate with pathogenic microbes and pollutants; D, mixing of contaminated leachate with surface water bodies pond and river; E, leaching of leachate to groundwater and contamination of drinking water bodies


Images of different landfill sites and treatment of landfill leachate using anaerobic processes
Selected Publications Related to Field:
-
Hemme, C., Green, S., Rishishwar, L., Prakash, O., Pettenato, A., Chakraborty, R., Deutchbauer, A., Van Nostrand, J., Wu, L., He, Z., Jordan, IK, Hazen, T., Adam Arkin, A., Kostka, J. and Zhou, J. (2016). Lateral Gene Transfer in a Heavy Metal- Contaminated Groundwater Microbial Community. 7(2):e02234-15. doi:10.1128/mBio.02234-15.
-
Jasrotia P, Green SJ, Canion A, Overholt W, Prakash O, Wafula D, Hubbard D, Watson D, Schadt C, Brooks S and Kostka JE (2014). Watershed scale fungal community characterization along a pH gradient in a subsurface environment co-contaminated with uranium and nitrate. Appl. Environ. Microbiol. 80:1810-1820 doi: 10.1128/AEM.03423-13
-
Prakash O and Lal R (2013). Role of unstable phenanthrene degrading Pseudomonas species in the natural attenuation of phenanthrene contaminated sites. Korean J Microbiol Biotechnol 41:79-87. ISSN: 1598642X
-
Kostka JE, Prakash O, Overholt W, Green S, Freyer G, Canion A, Delgardio J, Norton, N and Huettel M (2011). Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Appl. Environ. Microbiol. 77, 7962-7974
-
Green SJ, Prakash O, Akob DM, Gihring TM, Jardin P, Watson DB and Kostka JE (2010). Denitrifying bacteria isolated from terrestrial subsurface sediment exposed to mixed contamination. Appl Environ Microbiol 76, 3244-3254. (Equal authorship
-
Dadhwal M, Singh A, Prakash O, Gupta SK, Kumari K, Sharma P, Jit S, Verma M, Holliger C and Lal R (2009). Proposal of biostimulation for hexachlorocyclohexane (HCH)-decontamination and characterization of culturable bacterial community from high-dose point HCH-contaminated soils. J Appl Microbiol 106, 381-92. ISSN: 1365-2672
-
Gupta SK, Kumari R, Prakash O and Lal R (2008). Pseudomonas panipatensis sp. nov., isolated from an oil-contaminated site. Int J Syst Evol Microbiol 58, 1339-1345. ISSN 1466-5026
-
Jit S, Prakash O and Lal R (2008). Flavobacterium lindanitolerans sp. nov., isolated from hexachlorocyclohexane-contaminated soil, Int J Syst Evol Microbiol 58, 1665-1669. ISSN 1466-5026.
-
Raina V, Suar M, Singh A, Prakash O, Dadhwal M, Gupta SK and Lal R (2008). Enhanced biodegradation of hexachlorocyclohexane (HCH) in contaminated soil via inoculation of Sphingobium indicum B90A. Biodegradation 19, 27-40. ISSN: 0923-9820
-
Prakash O, Kumari K and Lal R (2007). Pseudomonas delhiensis sp. nov, from a fly ash dumping site of a thermal power plant. Int J Syst Evol Microbiol 57, 527-531. ISSN 1466-5026 (IF 2.8, 18).
-
Prakash O and Lal R (2006). Description of Sphingobium fuliginis sp. nov., a phenanthrene degrading bacterium from fly ash dumping site, and reclassification of Sphingomonas cloacae as Sphingobium cloacae comb. nov. Int J Syst Evol Microbiol 56, 2147-2152. ISSN 1466-5026 (IF 2.8, 18)
Study of Biomethanation and Methanotrophic Process in Rice Paddies and Landfills: Methane is the second most abundant greenhouse gas with >20 times more greenhouse potential than CO2. Methanogenic Archaea is the main producer (source) of biogenic methane, while methanotrophs consume a substantial part of generated methane and work as methane sinks. Landfill and rice paddies are potent biogenic methane sources and immensely contribute to the global carbon budget, warming, and climate change. India is the 2nd largest rice producer after China and has several unmanaged landfills nationwide. Both emit sufficient CH4, pose a risk for global warming and Climate change, and need immediate attention. At SCCCS, we are working on methanogenesis and methanotrophy and trying to understand the source and sink relationship of CH4 in rice paddies and landfills using geochemical, molecular and physiological approaches. Furthermore, data generated from the above process will assist in developing the right microbial-based mitigation strategies for the ongoing climate crisis.
Selected Publications Related to Field:
-
Sagar I, Nimonkar Y, Dhotre D, Shouche Y, Ranade D, Dewala S, Prakash, O* (2022). A microcosm model for the study of microbial community shift and carbon emission from different landfills of India. Ind J Microbiol. 62: 195 – 203.https://doi.org/10.1007/s12088-021-00995-7.
-
Nimonkar YS, Kajale S, Dake M, Ranade DR, Yadav KK, Kumar R, Prakash O. A culture-based and culture-independent approach to the study of landfill leachate bacterial and archaeal communities. Anaerobe. 2022 Aug 14:102626-.
-
Singh AK, Nakhate SP, Gupta RK, Chavan AR, Poddar BJ, Prakash O, Shouche YS, Purohit HJ, Khardenavis AA. Mining the landfill soil metagenome for denitrifying methanotrophic taxa and validation of methane oxidation in microcosm. Environmental Research. 2022 Dec 1;215:114199.
-
Cardzo S. and Prakash O. (2019). Landfill: Dump yard or Graveyard as featured article in Science Reporter.
-
Om Prakash and Kranti Karande (2020). Landfill leachate: A black devil for public and environmental health in Scientific Factor


View of open dumping sites and generated leachate. Due to heavy load of pollutants and pathogens untreated leachate has huge potential of soil and water contamination

Microbial Denitrification and Nitrous Oxide Release: Denitrification is anaerobic respiration in which microbes use NO3 as an alternative electron acceptor under anaerobic conditions without oxygen. Nitrate is a priority pollutant of water too. Heavy nitrate load induces eutrophication, hypoxia and fish death, while drinking nitrate-contaminated water creates several health problems. Denitrification plays an important role in wastewater treatment. Still, nitrous oxide released during denitrification works as a potent greenhouse gas and leads to the problem of global warming and climate change. As an agricultural economy, India uses immense amounts of nitrogenous fertilizers in farming practices. Being water-soluble, nitrate contaminates groundwater, soil, and surface water by leaching and surface runoff. Despite the agricultural-based economy, soil denitrification and nitrous oxide generation potential of Indian agricultural land are understudies. Using a geochemical molecular and physiological approach along with nitrate reduction, we are trying to understand the nitrous oxide release potential of different agroecosystems in India. We have cultivated >500 strains of denitrifying bacteria from soil, wastewater, and landfill leachate, working on their genomics, physiology and denitrification potentials.
Selected Publications Related to Field:
-
Canion A., Prakash O., Green S., Jahnke L, Kuypers M, Kostka J. (2013) Isolation and physiological characterization of psychrophilic denitrifying bacteria from permanently cold Arctic fjord sediments (Svalbard, Norway). Environ Microbiol. 15:1606-1618. doi:10.1111/1462-2920.12110
-
Venkatramanan R, Prakash O, Woyke T, Chain PSG, Goodwin LA, Watson DB, Brooks SC, Kostka JE and Green SJ (2013) Genome sequences for three denitrifying bacterial strains isolated from a uranium- and nitrate-contaminated subsurface environment. Genome Announcment. e00449-13. doi: 10.1128/genomeA.00449-13.
-
Prakash O, Green SJ, Jasrotia P, Overholt WA, Canion A, Watson DB, Jardine PM, Brooks SC and Kostka JE (2012). Rhodanobacter denitrificans sp. nov., isolated from nitrate-rich zones of a contaminated aquifer. Int J Syst Evol Microbiol 62: 2457–2462.
-
Green SJ, Prakash O, Jasrotia P, Overholt WA, Cardenas E, Hubbard D, Tiedje JM, Watson DB, Jardine PM, Brooks SC and Kostka JE (2012). Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site Appl. Environ. Microbiol.78: 1039-
-
Green SJ, Prakash O, Akob DM, Gihring TM, Jardin P, Watson DB and Kostka JE (2010). Denitrifying bacteria isolated from terrestrial subsurface sediment exposed to mixed contamination. Appl Environ Microbiol 76, 3244-3254.
Anaerobic Process of Clean Energy Generation from Residual Biomass: Biomass-based clean energy is one of the centre’s thrust areas. Methanogenic archaea and Clostridium species are key biomethane and biohydrogen production microorganisms, respectively. We are cultivating and characterizing efficient strains of Methanogens and Clostridium for biohydrogen, biobutanol and biomethane production from agricultural residues, Mandi waste, food waste and other biomass in collaboration with some industries involved in clean energy and BioCNG project and also working on the production of value-added fertilizers from sludge generated during this process.
Selected Publications Related to Field:
-
Sagar I, Nimonkar Y, Dhotre D, Shouche Y, Ranade D, Dewala S, Prakash, O* (2022). A microcosm model for the study of microbial community shift and carbon emission from different landfills of India. Ind J Microbiol. 62: 195 – 203.https://doi.org/10.1007/s12088-021-00995-7
-
Nimonkar YS, Kajale S, Dake M, Ranade DR, Yadav KK, Kumar R, Prakash O. A culture-based and culture-independent approach to the study of landfill leachate bacterial and archaeal communities. Anaerobe. 2022 Aug 14:102626-.
-
Singh AK, Nakhate SP, Gupta RK, Chavan AR, Poddar BJ, Prakash O, Shouche YS, Purohit HJ, Khardenavis AA. Mining the landfill soil metagenome for denitrifying methanotrophic taxa and validation of methane oxidation in microcosm. Environmental Research. 2022 Dec 1;215:114199.
-
Prakash, O., Dodsworth, J. A., Dong, X., Ferry, J. G., L’Haridon, S., Imachi, H., … & Whitman, W. B. (2023). Proposed minimal standards for description of methanogenic archaea. International Journal of Systematic and Evolutionary Microbiology, 73(4).

Demonstration of biogas formation from collected landfill leachate in serum vials microcosm. Gas chromatography data indicated 65-70% CH4 content in collected headspace gas
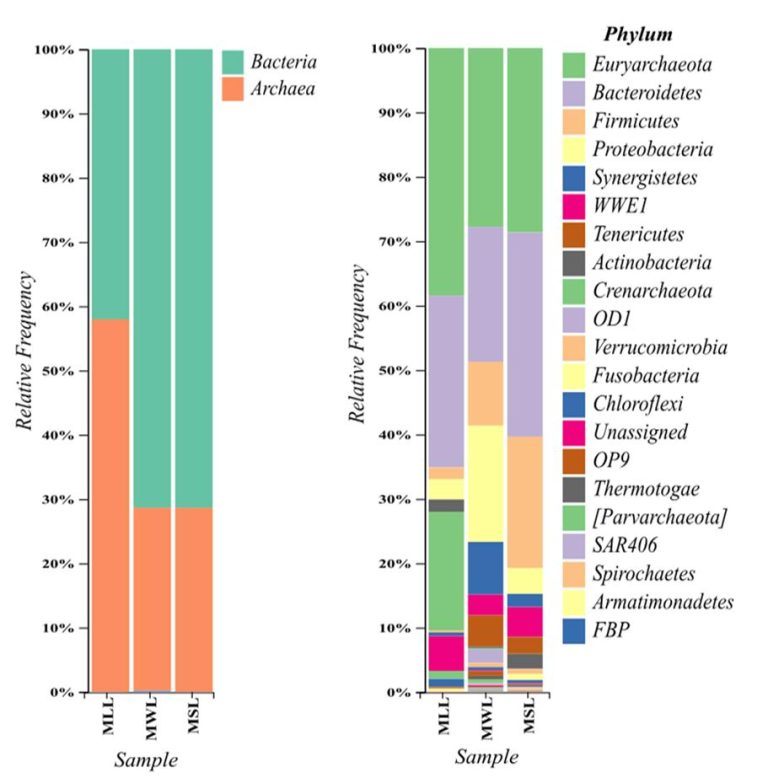
Metagenomic studies from three different landfill leachate sample indicate that Archaea constitute substantial part of leachate microbial community and play crucial role in ecology of landfill leachate.
Establishment of a Microbiome bank of unique Indian niches to study the impact of climate change on microbial diversity and ecosystem functionality: Despite immense work, to date, Microbiologists can only cultivate 1-10% microbial diversity from different ecosystems. Rest-90-99 % need to be-yet-cultured. It is proven that global warming, climate change, habitat destruction, urbanization and sudden entry of pollutants in the pristine ecosystem can lead to the extinction of certain microorganisms, which might be of immense clinical, industrial and biotechnological importance, and we can lose them without cultivation and exploitation. Along with developing the new media and technology for the cultivation of not-yet-cultured microbial taxa, we also focus on developing long-term intact microbiome preservation protocol. It will help to study the effect of climate change on microbial community structure and ecosystem functionality over time.
Selected Publications Related to Field:
-
Prakash O, Nimonkar Y, Desai D. (2020) A Recent Overview of Microbes and Microbiome Preservation. Ind J Microbiol. 60:297-309. doi: 10.1007/s12088-020-00880-9.
-
Prakash, O. and Jangid K. (2015). Preservation of uncultivated microbial cells for single cell genomics and cultivation in future. Single Cell Biology S1:003. doi:10.4172/2168-9431.S1-003
-
Prakash O, Shouche YS and Jangid K (2013). Microbial cultivation and the role of microbial resource centers in the omics era. Appl. Microbiol. Biotechnol. 53: 247-252 doi: 10.1007/s00253-012-4533-y
-
Prakash O, Nimonkar Y and Shouche YS (2013). Practice and prospects of microbial preservation. FEMS Microbiol Lett 339: 1-9 (doi:10.1111/1574-6968.12034)
-
Prakash O, Sharma A, Shouche YS (2020) Importance of Vision, Visibility and Vitality of Microbial Resource Centres. Curr. Sci. Current Science 119(4):625-631 DOI:10.18520/cs/v119/i4/625-631
Development of Archaeal bioinoculants for sustainable agriculture: Modern molecular tools have proven that archaea constitute a significant part of soil ecosystems. Most plant growth-promoting and biopesticide research mainly focused on bacterial and fungal bioinoculants. We are exploring the possibility of the role of archaea as bioinoculants for sustainable agriculture.
Selected Publications Related to Field:
-
Prakash O. (2022). Lack of kinship with anaerobes is a kind of short-sightedness of agricultural and environmental microbiologists. Environ. Microbiol. Rep. 14: 330-332. https://doi.org/10.1111/1758-2229.13058
-
Prakash O, Sharma A, Shouche YS (2020) Importance of Vision, Visibility and Vitality of Microbial Resource Centres. Curr. Sci. Current Science 119(4):625-631 DOI:10.18520/cs/v119/i4/625-631
-
Keswani C, Prakash O, Bharti N, Vílcheze JI, Sansinenea E, Lally RD, Borrissh R, Singh SP, Gupta VK, Fraceto LF, Limak R., Singh HB (2019). Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci Total Environ. 690:841-852
-
Bharti, N., Sharma, SK, Saini, S., Verma, A, Nimonkar, Y. and Prakash, O. (2017). Microbial Plant probiotics: Problems in application and formulation. In “Probiotics and Plant Health” pp. 317-335. Springer, Singapore.
-
Prakash, O Sharma, R, Singh, P., Yadav, A. (2015). Strategies for taxonomical characterization of agriculturally important microorganisms. In. Microbial Inoculants in Sustainable Agricultural Productivity. Springer India (DOI: 10.1007/978-81-322-2647-5_5).
High-throughput Cultivation, Microbial Taxonomy and Systematics: Cultivation permanently resides in the heart of microbiology. We have extended experience in cultivating and characterizing tough to cultivate Bacteria, Archaea and Fungi. We have published >15 novel taxa of bacteria, fungi and Archaea. We always haunt like a Gofer for the novel, not-yet-cultured and ecologically significant microbes to test the hypothesis generated from genome sequence data or metagenomics.
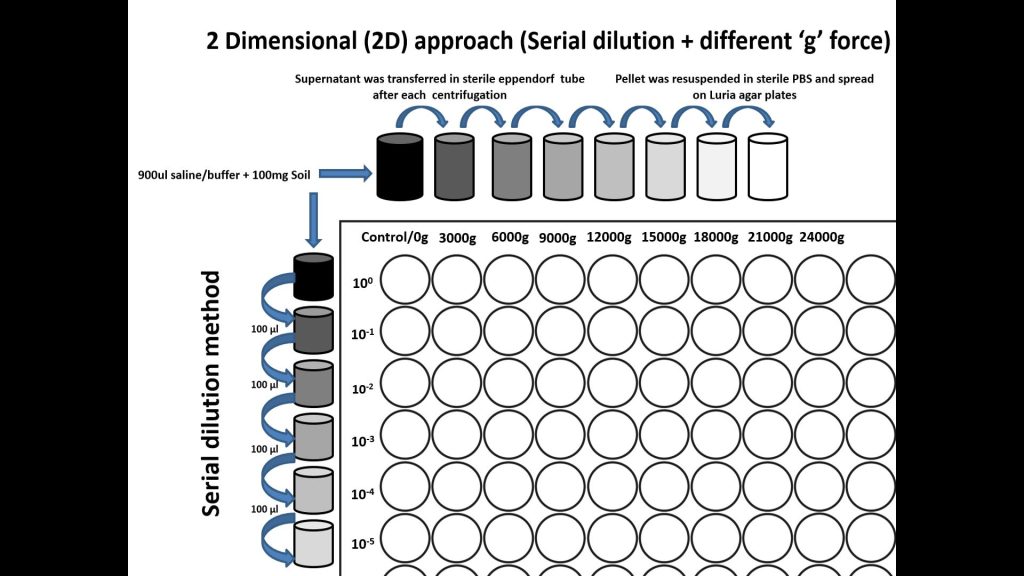
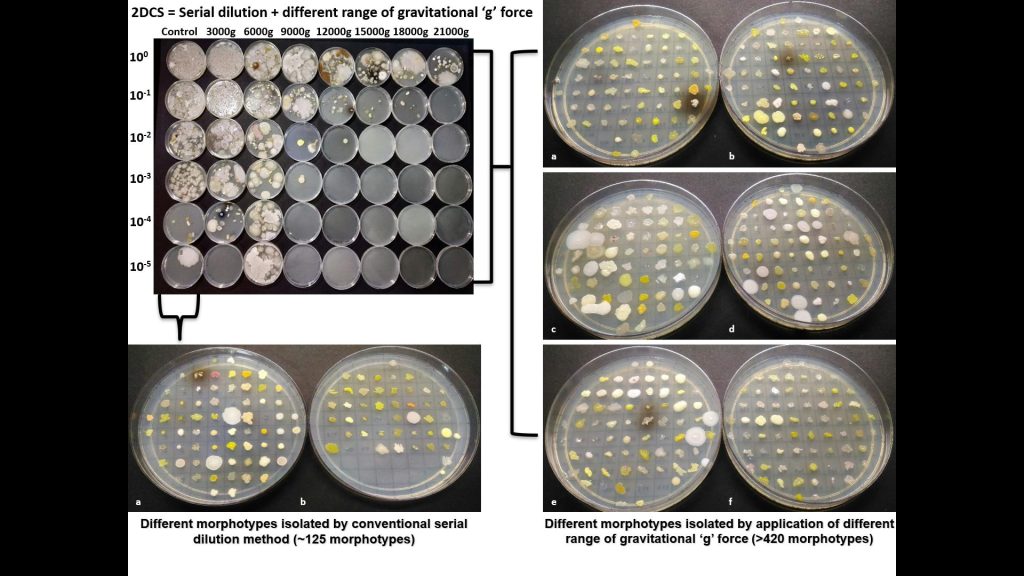
Concept of Two-dimensional cell separation a high -throughput approach of microbial cultivation.
Selected Publications Related to Field:
Yadav KK, Nimonkar Y., PoddarBJ, Kovali L., Sagar I1, Shouche Y, Purohit HJ, KhardenavisAA, Green SJ, Prakash O (2022). Two-Dimensional Cell Separation: A High Throughput Approach to Enhance the Culturability of Bacterial Cells from Environmental Samples. Microbiol. Spect. e00007-22. https://doi.org/10.1128/spectrum.00007-22 ISSN: 2165-0497
Nimonkar YS, Godambe T, Kulkarni A, Patel T, Paul D, Rale V, Prakash O. Oligotrophy vs. copiotrophy in an alkaline and saline habitat of Lonar Lake. Frontiers in Microbiology. 2022 Aug 4;13:939984-.
Joglekar A, Nimonkar Y, Bajaj A, Prakash O (2022) Resolution of inter/intraspecies variation in Weissella group requires multigene analysis and functional characterization . J. Basic Microbiol. https://doi.org/10.1002/jobm.202200357
*Prakash O, Parmar M, Vaijanapurkar M, Rale V, Shouche YS. Recent trend, biases, and limitations of cultivation based diversity studies of microbes. FEMS Microbiol Lett. 2021 368(17):fnab118. doi: 10.1093/femsle/fnab118..
Prakash O, Muduli S, Kumar R. Kumari C, Nimonkar Y, Shouche YS, Rakesh Sharma R. (2017). Description of Auricoccus indicus gen. nov., sp. nov., isolated from skin of human ear. Int J Syst Evol Microbiol. 67(5), 1212-1218
Sharma R., Prakash O, Sonawane MS, Nimonkar Y, Golellu P., Sharma R. (2016). Diversity and distribution of phenol oxidase producing fungi from Soda Lake and description of Curvularia lonarensis sp.nov. Front. Microbiol. 7: 1847. doi.org/10.3389/fmicb.2016.01847.
Prakash, O., Sharma, A., Nimonkar, Y., Shouche Y. S. (2015): Proposal for creation of a new genus Neomicrococcus gen. nov. for Zhihengliuella aestuarii and Micrococcus lactis (Chittpurna et al., 2011). Int J Syst Evol Microbiol 65(11), 3771-3776 doi: 10.1099/ijsem.0.000490.
Prakash, O., Nimonkar, Y., Vaishampayan, A., Mishra, M., Kumbhare, S., Josef, N., Yogesh Shouche, YS, (2015). Description of Pantoea intestinalis sp. nov. : isolated from Human gut.” Int. J. Syst. Evol Microbiol 65(10), 3352-3358.doi: 10.1099/ijsem.0.000419
Prakash O, Munot H, Nimonkar Y, Sharma M, Kumbhare S, Shouche YS (2014) Description of Pelistega indica isolated from Human gut. Int J Syst Evol Microbiol 64: 1389-1394 doi: 10.1099/ijs.0.059782-0.
Prakash O, Green SJ, Jasrotia P, Overholt WA, Canion A, Watson DB, Jardine PM, Brooks SC and Kostka JE (2012). Rhodanobacter denitrificans sp. nov., isolated from nitrate-rich zones of a contaminated aquifer. Int J Syst Evol Microbiol 62: 2457–2462.
Prakash O, Kumari K and Lal R (2007). Pseudomonas delhiensis sp. nov, from a fly ash dumping site of a thermal power plant. Int J Syst Evol Microbiol 57, 527-531.
Prakash O and Lal R (2006). Description of Sphingobium fuliginis sp. nov., a phenanthrene degrading bacterium from fly ash dumping site, and reclassification of Sphingomonas cloacae as Sphingobium cloacae comb. nov. Int J Syst Evol Microbiol 56, 2147-2152.
Cultivation, characterization and biobanking of obligate anaerobes of ecological significance: Anaerobes play a significant role in various ecosystem services like greenhouse gas generation, metal reduction, solid waste management and human-gut ecology. We are cultivating characterizing and biobanking Methanogens, sulphate reducers, metal reducers, denitrifiers and obligate anaerobes with various metabolism. The human gut, anaerobic infection sites, anaerobic digester, landfill leachate and sediments are some of the target areas for our cultivation.


Demonstration of pre-reduced medium preparation, Hungate-tube incubation and Roll-Tube isolation of obligate anaerobes

Methodological demonstration of serum vials enrichment, detection under fluorescent microscope and isolation of methanogens
Selected Publications Related to Field:
-
Nimonkar, Y. S., S. Kajale, M. Dake, D. R. Ranade, K. K. Yadav, R. Kumar, and O. Prakash. “A culture-based and culture-independent approach to the study of landfill leachate bacterial and archaeal communities.” Anaerobe (2022): 102626-102626.
-
Comparison of taxonomic resolutions of various typing methods for Clostridium and Paraclostridium Species Isolated from Landfill Leachate”, which you submitted to Current Microbiology.
-
Prakash O, Gihring TM, Dalton DD, Chin KJ, Green SJ, Akob DM, Wanger G and Kostka JE (2010). Geobacter daltonii sp. nov., an Fe(III)- and uranium(VI)-reducing bacterium isolated from the shallow subsurface exposed to mixed heavy metal and hydrocarbon contamination. Int J Syst Evol Microbiol 60, 546-553.
Roll tube anaerobic chamber
Role of landfill and landfill leachate in spreading pollutants and pathogens: Landfill work as open dumping sites for discarded materials. Due to ideal moisture conditions, sufficient organic loads and temperatures, it works as a breeding ground for different pathogens. The black and brown colour leachate generated from landfill sites contains a heavy load of pollutants and pathogens, and it disseminates them in freshwater and soil by the process of recharge, discharge and irrigation. We are also looking at how landfill leachate plays a role in the spread of pollutants and pathogens in the natural ecosystem and how to develop a method for landfill management.
Cycle and paper
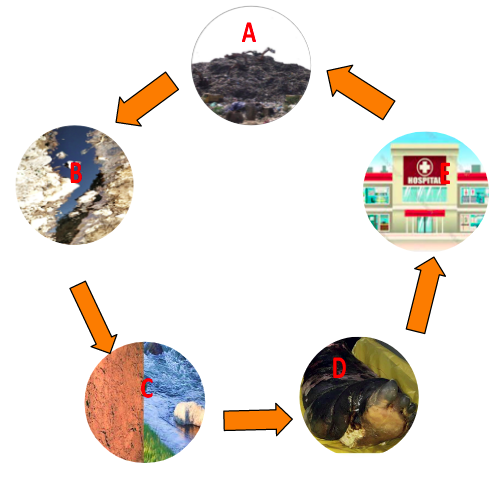
Graphical Representation of the role of landfill (A) and generated landfill leachate (B) in soil and water contamination (C) and community infection (D)
Residual Antibiotics and Their Role in the Development of Antimicrobial Resistance: Due to the indiscriminate use of antibiotics, India release a massive load of residual antibiotics in the natural environments, including sewage, soil, and water. We are trying to understand what are the relative residual concentration of antibiotics in nature and how they are affecting drug tolerance, microbial community structure and in development of AMR in natural microbial community
Selected Publications Related to Field:
-
Kovale L, Nimonkar Y, Green SJ, Shouche YS, Prakash O. Antibiotic susceptibility of human gut-derived facultative anaerobic bacteria is different under aerobic versus anaerobic test conditions. Microbes Infect. 2021 Jun 8:104847. doi: 10.1016/j.micinf.2021.104847.
-
Marano RBM, Fernandes T, A global multinational survey of cefotaxime-resistant coliforms in urban wastewater treatment plants. Environ Int. 2020.144:106035. doi: 10.1016/j.envint.2020.106035.
-
Nimonkar, Y.S., Yadav, B., Talreja, P. Prakash O (2019). Assessment of the Role of Wastewater Treatment Plant in Spread of Antibiotic Resistance and Bacterial Pathogens Ind J Microbiol. 59: 261–265 https://doi.org/10.1007/s12088-019-00793-2e


